REDCapTidieR
Making analysts lives easier through the power of tidy tibbles
8/18/22
Who am I?

- CGT DataOps Data Scientist
- 5 years at CHOP
- Engineering background
- Trying to implement
quartotoday1
R / Medicine 2022

https://events.linuxfoundation.org/r-medicine/
R / Medicine 2022 - Workshops

Discount Code: RMED22CHOP for 10% off!
R / Medicine 2022 - Speaker Highlights
- Stephan Kadauke (CHOP CGT DataOps)
R/Medicine 101: Intro to R for Clinical Data
Should we Teach Data Science to Physicians-in-Training?
- Joy Payton (CHOP Arcus Education)
- Using Public Data and Maps for Powerful Data Visualizations
- Lihai Song (CHOP Data Scientist)
- Automation of statistics summary and analysis using R Shiny
- Jaclyn Janis (RStudio/Posit, CHOP Representative)
- It’s time for nurses to learn R
Agenda
In today’s talk we will:
- Review what REDCap is 💡
- Review
REDCapRas an extraction tool for the API 🔌 - Implement
REDCapTidieRto make our lives easier 🧹
What you need:
- Familiarity with R 💻
- Familiarity with REDCap 🧢
What is REDCap?

- Free1 database solution for research
- Secure and accessible from a web browser
- Can collect “any type of data in any environment”
- Particularly useful for compliance with 21 CFR Part 11, HIPAA, etc.
- Requires little to get up and running, but offers complexity as needed
What is REDCap?
Record Status Dashboard 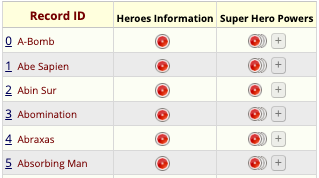
Front-End Data Entry UI 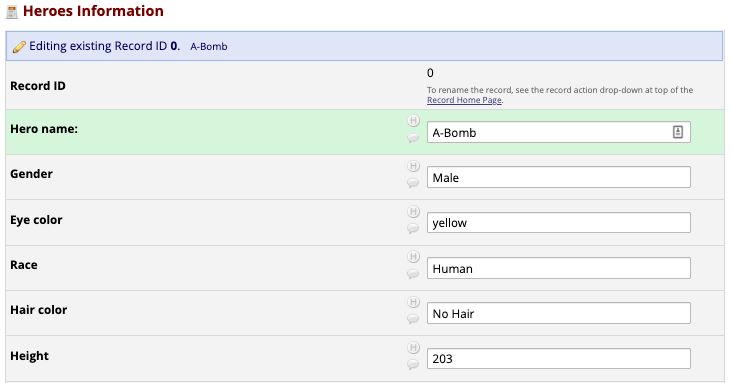
REDCap functions as a large data table, but data distribution can be complex depending on architectural choices.
Repeating instances can create headaches on the backend.
The Super Heroes Dataset
Open source dataset from SuperHeroDB and available on Kaggle. It contains two tables:
- Super Hero Information (i.e. demographic data)
- Super Hero Powers (i.e.
TRUE/FALSEfor specific powers)

On the Shoulders of Giants
Some core REDCapR functions:
redcap_read_oneshotredcap_metadata_readredcap_event_instruments- New as of
v1.1.0
- New as of
Requirements:
- Active REDCap project
- A REDCap API URI1
- API token2
SuperHeroes Output
# Load applicable libraries:
library(dplyr)
library(REDCapR)
superheroes_db <- redcap_read_oneshot(redcap_uri, token, verbose = FALSE)$data
superheroes_db %>%
glimpse()Rows: 6,700
Columns: 16
$ record_id <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, …
$ redcap_repeat_instrument <chr> NA, "super_hero_powers", "super_hero_power…
$ redcap_repeat_instance <dbl> NA, 1, 2, 3, 4, 5, 6, 7, NA, 1, 2, 3, 4, 5…
$ name <chr> "A-Bomb", NA, NA, NA, NA, NA, NA, NA, "Abe…
$ gender <chr> "Male", NA, NA, NA, NA, NA, NA, NA, "Male"…
$ eye_color <chr> "yellow", NA, NA, NA, NA, NA, NA, NA, "blu…
$ race <chr> "Human", NA, NA, NA, NA, NA, NA, NA, "Icth…
$ hair_color <chr> "No Hair", NA, NA, NA, NA, NA, NA, NA, "No…
$ height <dbl> 203, NA, NA, NA, NA, NA, NA, NA, 191, NA, …
$ weight <dbl> 441, NA, NA, NA, NA, NA, NA, NA, 65, NA, N…
$ publisher <chr> "Marvel Comics", NA, NA, NA, NA, NA, NA, N…
$ skin_color <chr> "-", NA, NA, NA, NA, NA, NA, NA, "blue", N…
$ alignment <chr> "good", NA, NA, NA, NA, NA, NA, NA, "good"…
$ heroes_information_complete <dbl> 0, NA, NA, NA, NA, NA, NA, NA, 0, NA, NA, …
$ power <chr> NA, "Accelerated Healing", "Durability", "…
$ super_hero_powers_complete <dbl> NA, 0, 0, 0, 0, 0, 0, 0, NA, 0, 0, 0, 0, 0…Remember redcap_repeat_instrument and redcap_repeat_instance, they’re coming back!
SuperHeroes Output
| record_id | redcap_repeat_instrument | redcap_repeat_instance | name | gender | eye_color | race | hair_color | height | weight | publisher | skin_color | alignment | heroes_information_complete | power | super_hero_powers_complete |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | NA | NA | A-Bomb | Male | yellow | Human | No Hair | 203 | 441 | Marvel Comics | - | good | 0 | NA | NA |
| 0 | super_hero_powers | 1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | Accelerated Healing | 0 |
| 0 | super_hero_powers | 2 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | Durability | 0 |
| 0 | super_hero_powers | 3 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | Longevity | 0 |
| 0 | super_hero_powers | 4 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | Super Strength | 0 |
| 0 | super_hero_powers | 5 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | Stamina | 0 |
| 0 | super_hero_powers | 6 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | Camouflage | 0 |
| 0 | super_hero_powers | 7 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | Self-Sustenance | 0 |
| 1 | NA | NA | Abe Sapien | Male | blue | Icthyo Sapien | No Hair | 191 | 65 | Dark Horse Comics | blue | good | 0 | NA | NA |
| 1 | super_hero_powers | 1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | Agility | 0 |
REDCap Repeating Instruments
Record Status Dashboard 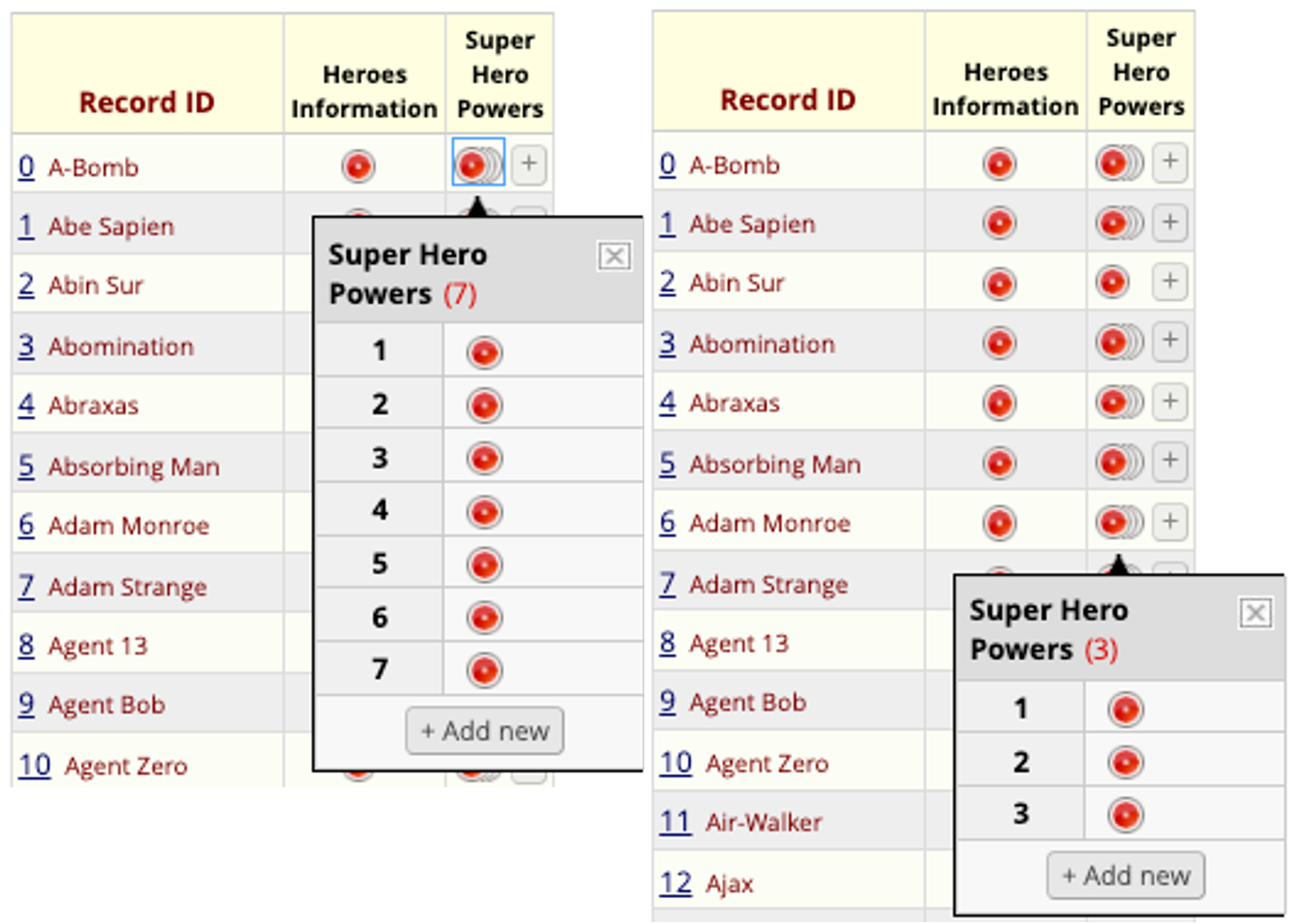
Front-End Data Entry UI 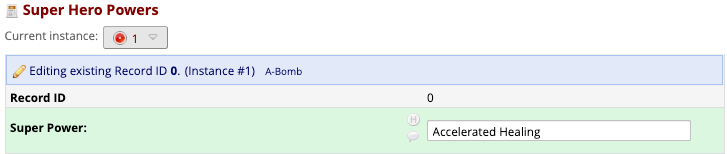
SuperHeroes Repeating Output
| record_id | redcap_repeat_instrument | redcap_repeat_instance | name | power |
|---|---|---|---|---|
| 0 | NA | NA | A-Bomb | NA |
| 0 | super_hero_powers | 1 | NA | Accelerated Healing |
| 0 | super_hero_powers | 2 | NA | Durability |
| 0 | super_hero_powers | 3 | NA | Longevity |
| 0 | super_hero_powers | 4 | NA | Super Strength |
| 0 | super_hero_powers | 5 | NA | Stamina |
| 0 | super_hero_powers | 6 | NA | Camouflage |
| 0 | super_hero_powers | 7 | NA | Self-Sustenance |
record_id,redcap_repeat_instrument, andrecap_repeat_instanceform a compound key.
A compound key is the combination of 2+ columns necessary to identify a row uniquely in a table
The Problem
- Empty data introduced as an artifact of repeating instruments
- Data export is often times large and unwieldy
- Missing metadata linking field association to instruments
- Row identification is confusing and inconsistent

Introducing REDCapTidieR
At a glance:
- Built on top of
REDCapR - Takes two inputs: REDCap URI and REDCap API token
- Returns a set of tidy
tibbles- One for each REDCap instrument
Revisiting Superheroes
# A tibble: 2 × 10
redcap_form_name redcap_form_label redcap_data redcap_metadata structure
<chr> <chr> <list> <list> <chr>
1 heroes_information Heroes Information <tibble> <tibble [11 × 17]> nonrepea…
2 super_hero_powers Super Hero Powers <tibble> <tibble [2 × 17]> repeating
# ℹ 5 more variables: data_rows <int>, data_cols <int>, data_size <lbstr_by>,
# data_na_pct <formttbl>, form_complete_pct <formttbl>Revisiting Superheroes
Non-Repeating Hero Information
| record_id | redcap_form_instance | power | form_status_complete |
|---|---|---|---|
| 0 | 1 | Accelerated Healing | Incomplete |
| 0 | 2 | Durability | Incomplete |
| 0 | 3 | Longevity | Incomplete |
| 0 | 4 | Super Strength | Incomplete |
| 0 | 5 | Stamina | Incomplete |
| 0 | 6 | Camouflage | Incomplete |
| 0 | 7 | Self-Sustenance | Incomplete |
| 1 | 1 | Agility | Incomplete |
| 1 | 2 | Accelerated Healing | Incomplete |
| 1 | 3 | Cold Resistance | Incomplete |
Repeating Hero Powers
| record_id | name | gender | eye_color | race | hair_color | height | weight | publisher | skin_color | alignment | form_status_complete |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | A-Bomb | Male | yellow | Human | No Hair | 203 | 441 | Marvel Comics | - | good | Incomplete |
| 1 | Abe Sapien | Male | blue | Icthyo Sapien | No Hair | 191 | 65 | Dark Horse Comics | blue | good | Incomplete |
| 2 | Abin Sur | Male | blue | Ungaran | No Hair | 185 | 90 | DC Comics | red | good | Incomplete |
| 3 | Abomination | Male | green | Human / Radiation | No Hair | 203 | 441 | Marvel Comics | - | bad | Incomplete |
| 4 | Abraxas | Male | blue | Cosmic Entity | Black | -99 | -99 | Marvel Comics | - | bad | Incomplete |
| 5 | Absorbing Man | Male | blue | Human | No Hair | 193 | 122 | Marvel Comics | - | bad | Incomplete |
| 6 | Adam Monroe | Male | blue | - | Blond | -99 | -99 | NBC - Heroes | - | good | Incomplete |
| 7 | Adam Strange | Male | blue | Human | Blond | 185 | 88 | DC Comics | - | good | Incomplete |
| 8 | Agent 13 | Female | blue | - | Blond | 173 | 61 | Marvel Comics | - | good | Incomplete |
| 9 | Agent Bob | Male | brown | Human | Brown | 178 | 81 | Marvel Comics | - | good | Incomplete |
Revisiting Superheroes
Non-Repeating Hero Information
| record_id | --- | form_status_complete |
|---|---|---|
| 0 | ... | Incomplete |
| 0 | ... | Incomplete |
| 0 | ... | Incomplete |
| 0 | ... | Incomplete |
| 0 | ... | Incomplete |
Change in
*_form_status_completetoform_status_complete
The Default Output
# A tibble: 2 × 10
redcap_form_name redcap_form_label redcap_data redcap_metadata structure
<chr> <chr> <list> <list> <chr>
1 heroes_information Heroes Information <tibble> <tibble [11 × 17]> nonrepea…
2 super_hero_powers Super Hero Powers <tibble> <tibble [2 × 17]> repeating
# ℹ 5 more variables: data_rows <int>, data_cols <int>, data_size <lbstr_by>,
# data_na_pct <formttbl>, form_complete_pct <formttbl>bind_tables Direct to Environment
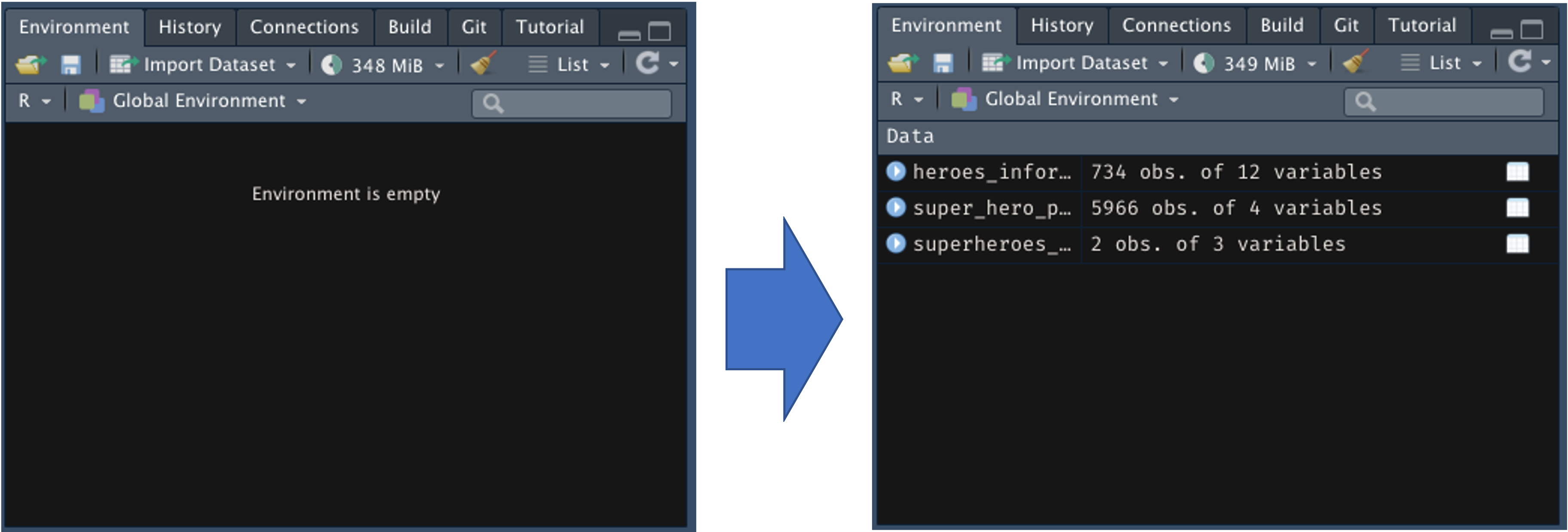
The function:
Clear out our envionrment:
Empty output, no global environment objects
Reload the superheroes_tidy dataset, pipe to bind_tables, check environment:
bind_tables Direct to Environment
Longitudinal REDCap Projects
| Classic | Longitudinal, one arm |
Longitudinal, multi-arm |
|
|---|---|---|---|
| Nonrepeated | record_id |
record_id +redcap_event |
record_id +redcap_event +redcap_arm |
| Repeated | record_id +redcap_repeat_instance |
record_id +redcap_repeat_instance +redcap_event |
record_id +redcap_repeat_instance +redcap_event +redcap_arm |
REDCap Projects with Arms
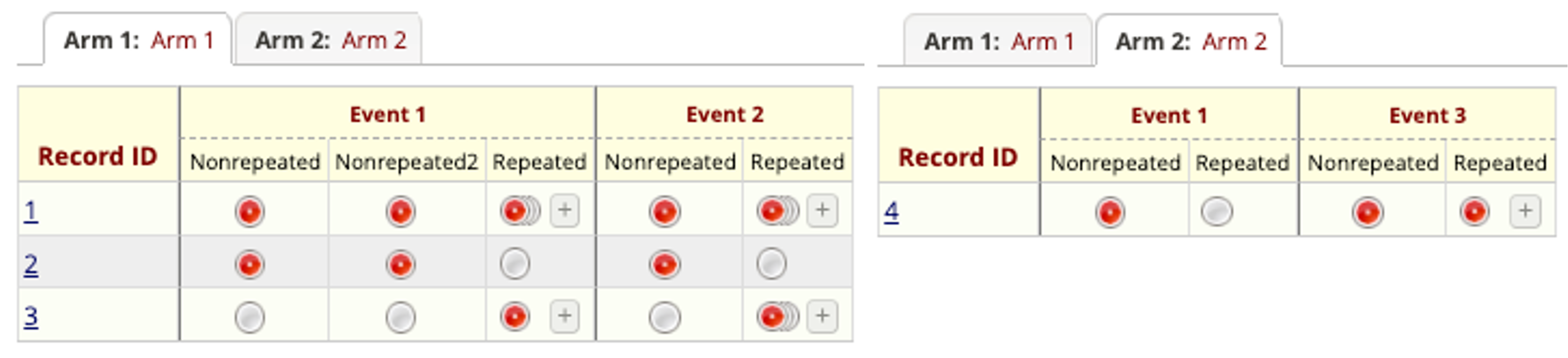
# A tibble: 3 × 11
redcap_form_name redcap_form_label redcap_data redcap_metadata redcap_events
<chr> <chr> <list> <list> <list>
1 nonrepeated Nonrepeated <tibble> <tibble [3 × 17]> <tibble>
2 nonrepeated2 Nonrepeated2 <tibble> <tibble [3 × 17]> <tibble>
3 repeated Repeated <tibble> <tibble [3 × 17]> <tibble>
# ℹ 6 more variables: structure <chr>, data_rows <int>, data_cols <int>,
# data_size <lbstr_by>, data_na_pct <formttbl>, form_complete_pct <formttbl>REDCap Projects with Arms
| record_id | redcap_event | redcap_arm | nonrepeat_1 | nonrepeat_2 | form_status_complete |
|---|---|---|---|---|---|
| 1 | event_1 | 1 | 1 | 2 | Incomplete |
| 1 | event_2 | 1 | 1 | 2 | Incomplete |
| 2 | event_1 | 1 | A | B | Incomplete |
| 2 | event_2 | 1 | 5 | 6 | Incomplete |
| 4 | event_1 | 2 | Red | Blue | Incomplete |
| 4 | event_3 | 2 | Green | Yellow | Incomplete |
| record_id | redcap_event | redcap_arm | redcap_form_instance | repeat_1 | repeat_2 | form_status_complete |
|---|---|---|---|---|---|---|
| 1 | event_1 | 1 | 1 | 1 | 2 | Incomplete |
| 1 | event_1 | 1 | 2 | 3 | 4 | Incomplete |
| 1 | event_1 | 1 | 3 | 5 | 6 | Incomplete |
| 1 | event_2 | 1 | 1 | A | B | Incomplete |
| 1 | event_2 | 1 | 2 | C | D | Incomplete |
| 3 | event_1 | 1 | 1 | C | D | Incomplete |
| 3 | event_2 | 1 | 1 | E | F | Incomplete |
| 3 | event_2 | 1 | 2 | G | H | Incomplete |
| 4 | event_3 | 2 | 1 | R1 | R2 | Incomplete |
Try it for Yourself!*
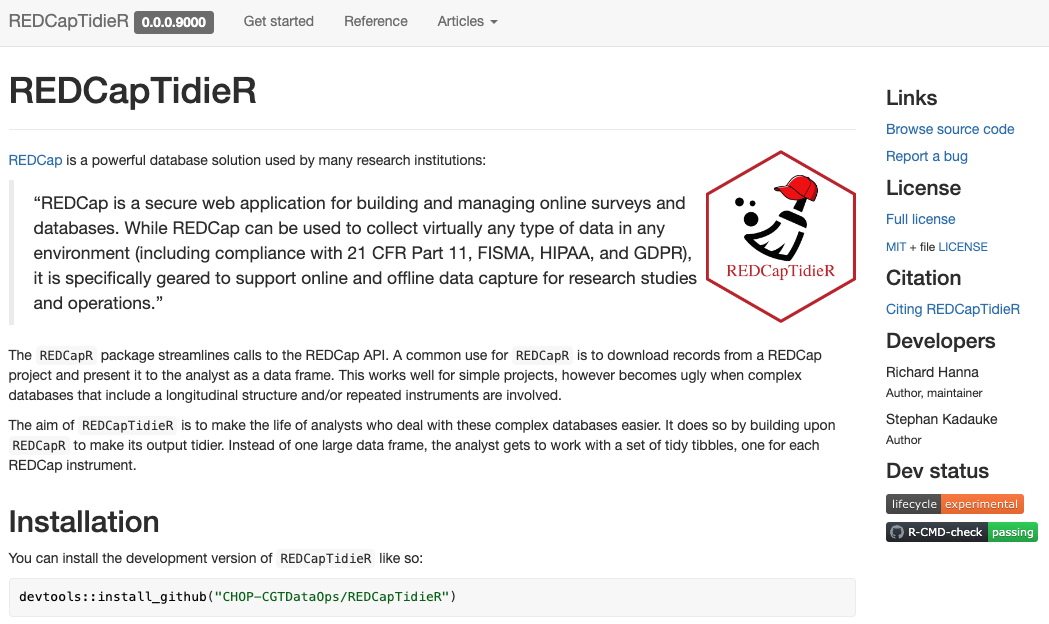
Install from public GitHub and view the pkgdown site
*REDCapTidieR is in early alpha, but hopefully for not for long!
Future Work
raw_or_labelcompatibilityextract_tableandextract_tablesfunctions- Release to CRAN
REDCap Metadata
metadata <- redcap_metadata_read(redcap_uri, token)$data
metadata %>%
kbl(booktabs = T, escape = F, table.attr = "style='width:20%;'") %>%
# options for HTML output
kable_styling(bootstrap_options = c("striped", "hover", "bordered"),
position = "center",
full_width = F,
font_size = 12,
fixed_thead = T) %>%
column_spec(1, bold = T) %>%
scroll_box(width = "100%", height = "300px")